
Quantitative x-ray computed tomography is used for structural and chemical analysis. After an instrument has been well calibrated, it is possible to recover the concentration of hydroxyapatite in samples of bone. This can be valuable information for monitoring bone growth and recovery, especially in experimental settings. Further, the high-resolution structural data can be used to gather morphological parameters that are indicators of structural integrity and healthy growth. In this study, rabbits were implanted with a set of four growth scaffolds connecting the transverse processes of the spine with the goal of local bone growth. The implants were monitored longitudinally in vivo by scanning longitudinally over several weeks post implantation. In addition, some implants were resected ex vivo and imaged using a micro CT.
Figure 1
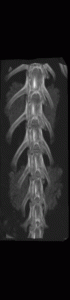
However, as these implants continued their growth, they became difficult to distinguish from the bones onto which they were grafted. This, in turn, made the quantitative structural and chemical analysis prohibitively difficult. To segment these bony implants with higher fidelity, a scheme was developed to first remove static bones in order to more readily isolate the implants. Individual vertebrae are isolated in the scan from the earliest time point (Figure 2).
Figure 2
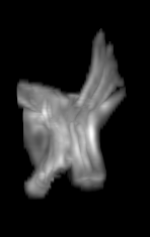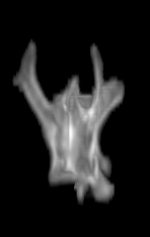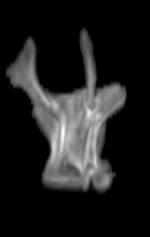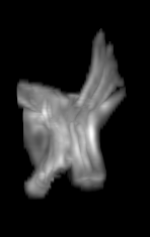
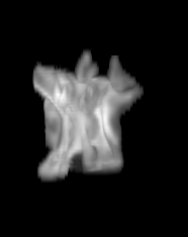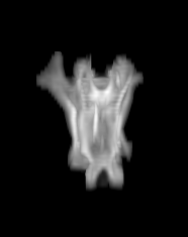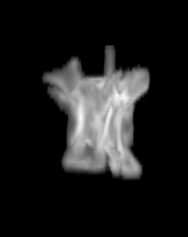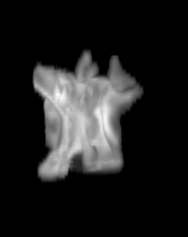
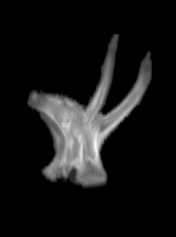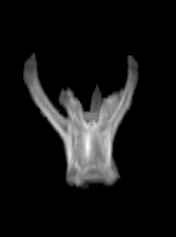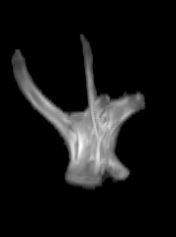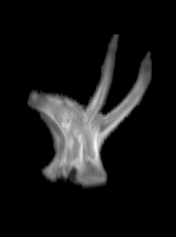
For each subject, each vertebra was isolated at the earliest available time point, where the bone growth in the implants was not yet sufficient to corrupt the local bone signal. These vertebrae were then rigidly co-registered by subject to all subsequent time points. The entirety of each vertebral region was then cut from the image, allowing for a straightforward segmentation of each implant. This not only increased the ease of segmentation, but also the accuracy since the native bone was unable to corrupt higher level analysis.
Figure 3
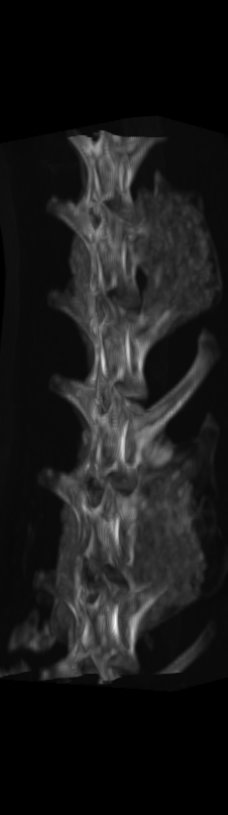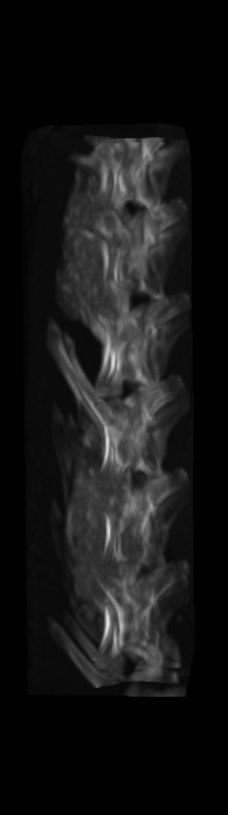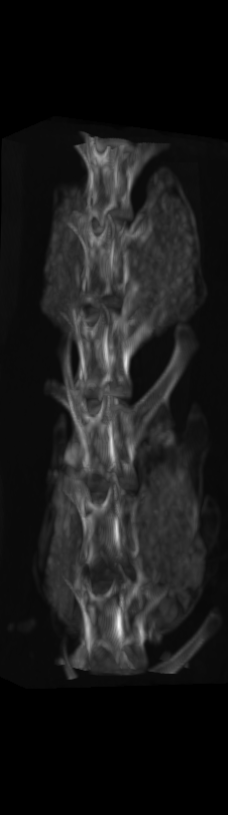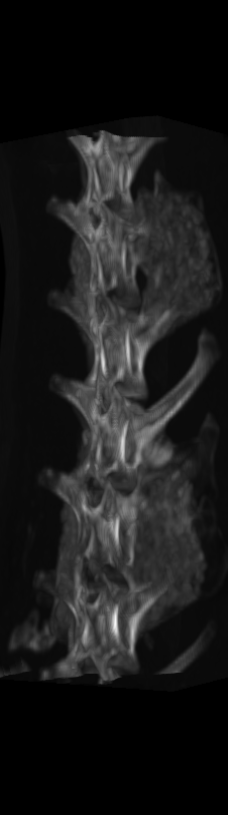
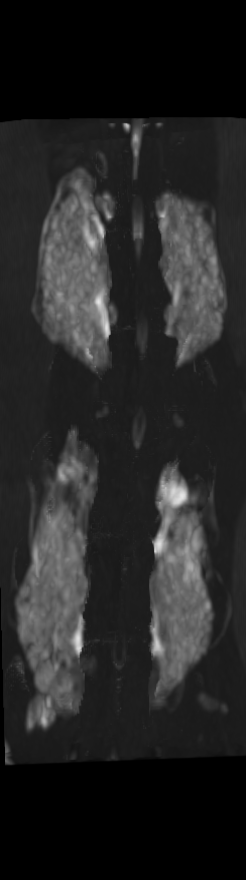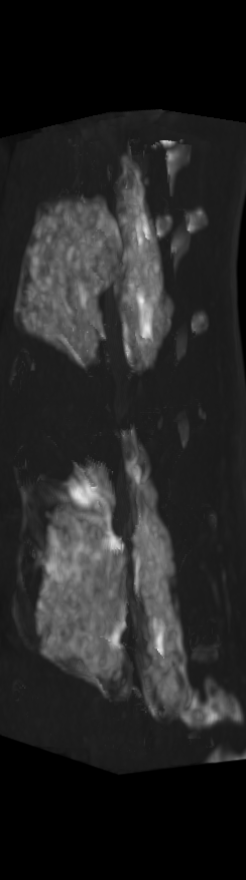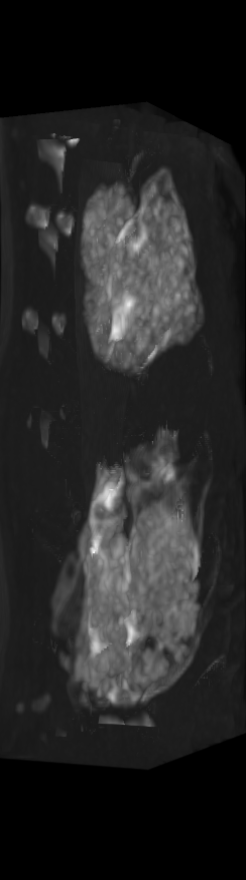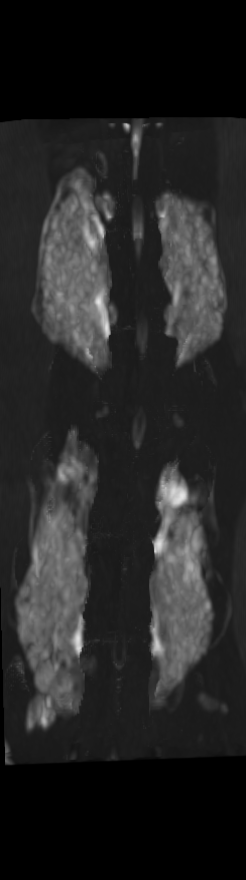

A) For later time points, the implants can be difficult to isolate from the bone.
B) The vertebrae from earlier time points are registered and removed.
C) The implants are easily segmented. The final segmentation of the implants is robust, and the image is prepared for higher level analysis.
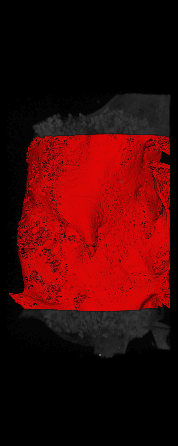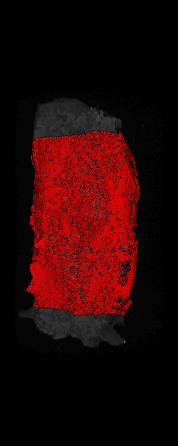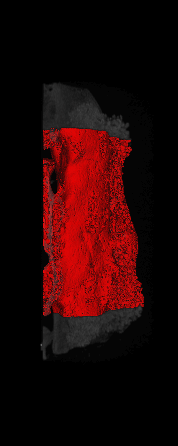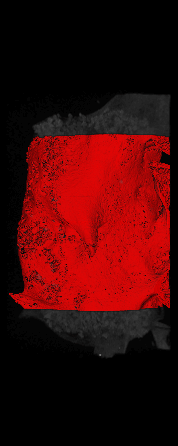
The ex vivo samples that were measured using high resolution x-ray CT were then analyzed for their structural integrity. In principle it is possible to construct a reliable finite elements model to predict strength under loads. However, these models require a prohibitive amount of training and computational footprint for high resolution samples. Instead, a structural parameter based on the electrical analogue of conductivity was used to analyze the samples for structure. First, the samples were segmented from the background using a two-call, one threshold entropy minimization histogram method. Due to biological variation, each sample was a slightly different length. To accurately compare samples to each other, it was also necessary to isolate a uniform length. Finally, an adaptive, dual-direction morphological connectivity scheme was used to isolate the subsections of the implant which were structurally relevant given a simulated end load (Figure 4).
Figure 4
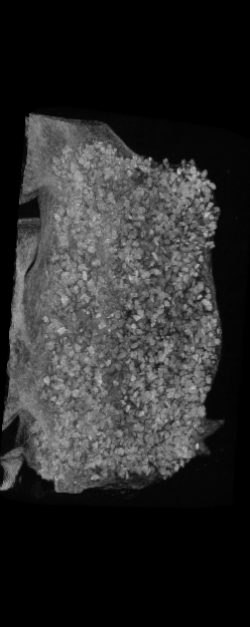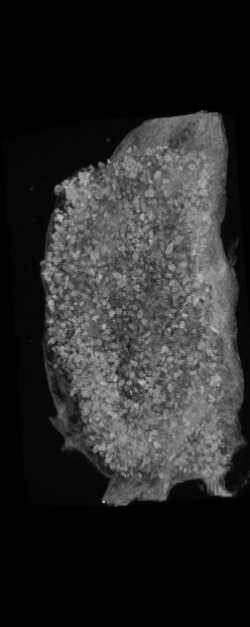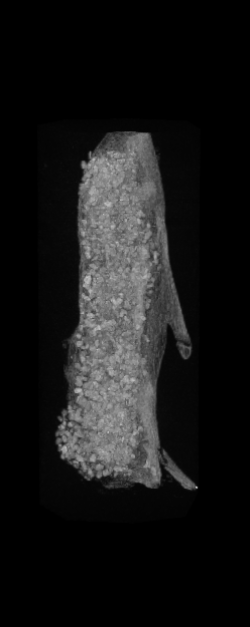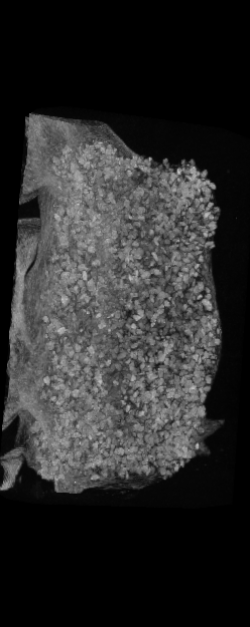
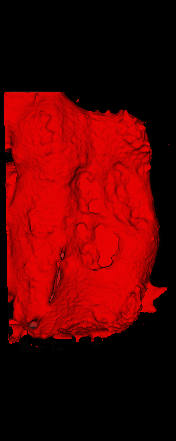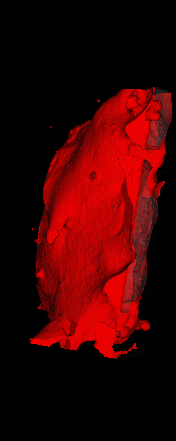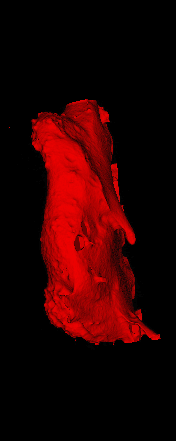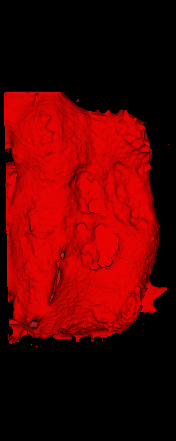

A) Original microCT Image
B) Implant Segmentation
C) Crop to Common Space for Uniform Comparison Across Subjects
D) Morphological Processing to Isolate Structural Components
The longitudinal segmentation scheme provided notable decreases in processing time and increases in accuracy with respect to hand segmentation. This is one example of many of the development and use of nonstandard but creative segmentation and quantification methods at Invicro.
